Home › Forums › FABRIC General Questions and Discussion › File save error and Load file error
- This topic has 7 replies, 4 voices, and was last updated 3 years, 1 month ago by
Ilya Baldin.
-
AuthorPosts
-
January 30, 2023 at 5:06 pm #3709
Hello,
We were running a layer 2 network test experiment with our Jupyter notebook. We created the notebook within JupyterHub. After we requested the slice, a file save error popped up. Our network works properly. Please see the detail information below:
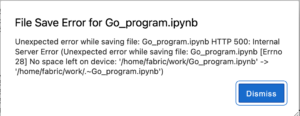
While the same time, we could not open other example notebooks neither. The error shows that file load error for example.ipynb. Please see the screenshot below:

I am not sure what might cause this happen. I was wondering if I could have some help with this issue.
Thanks,
Xusheng-
This topic was modified 3 years, 1 month ago by
Xusheng Ai.
-
This topic was modified 3 years, 1 month ago by
Xusheng Ai.
January 30, 2023 at 5:21 pm #3714It looks like you filled your disk allocation our JupyterHub. Do you have old files that you can clean up?
January 30, 2023 at 5:31 pm #3724I delete some old files, and it works well right now. Thank you so much for the hints.
January 30, 2023 at 5:32 pm #3725Is there any chance that we could have a larger disk on JupyterHub?
January 30, 2023 at 5:35 pm #3726What are you using it for? Generally, the JupyterHub is a good place for code/script/docs (i.e. smaller things). Do you need space for large data sets? If so we can create a persistent storage volume in the testbed itself.
January 30, 2023 at 6:06 pm #3727We are trying to upload one 1G bio-related file to the slice nodes through JupyterHub. Since there is no way to upload files from local to Fabric nodes directly. I checked the disk space that we have 1G available disk space in total. If it was possible to create a persistent storage volume in the testbed, it will be much appreciated.
January 30, 2023 at 6:20 pm #3728there is no way to upload files from local to Fabric nodes directly
It’s possible in two ways:
- Host your file with an HTTPS server somewhere on the Internet (with HTTP Basic authentication if desired), and download it on the nodes with wget command.
- Add the nodes into your local ~/.ssh/config with ProxyJump through the bastion, and then run scp to upload the file to the nodes.
I’ve done both in different experiments, but only the first one can be automated.
January 31, 2023 at 6:07 pm #3766Just as a form of explanation – we host the Jupyter Hub in Google Cloud, which costs real $$s allocated to us from NSF via a project named CloudBank. We are still evaluating the true costs of running it in its current configuration (so we can more accurately project future costs). We may revise the amount of disk space and other resources each notebook server gets, however we are constrained by the budget and this will not be a decision we will be making in the near term.
In general the Hub is not intended as a place to park or transfer large files.
-
This topic was modified 3 years, 1 month ago by
-
AuthorPosts
- You must be logged in to reply to this topic.
