Forum Replies Created
-
AuthorPosts
-
@Xusheng Manas is likely correct.
The FABRIC development team ran a load test this morning to test scaling of the framework (i.e. having 100s of people all try to submit slices at the same time). This broke/slowed some things throughout the day. Hopefully, by end-of-day Sunday, FABRIC will be improved with the changes mentioned in the post and be back to being stable.
Edit – I forgot to refresh the page so I didn’t see your reply @Chengyi
That’s great, I’m glad you got it working! None of the other sites I’ve tested are close to 90 Gbps, most are around 10 Gbps. Maybe the numa domains issue @Paul suggested.
– Brandon
Nevermind, just re-ran the tests again, now at off-hours and got:
SALT -> UTAH: 91.98 Gbps
UTAH -> SALT: 96.938 GbpsSo it must have been users or something.
Okay, the plot thickens!
I just ran more tests and I just achieved 98.061 Gbps from UTAH -> SALT (with 32 parallel streams), but only 30.653 Gbps from SALT -> UTAH. (The one thing I fixed to get back to 97 Gbps was I accidentally had turned off fair-queuing before.)
Time to put on our thinking caps!
@Chengyi, yes I’ll see if I can upload my notebook, but in case I can’t, here are the tuning parameters:
I’m using ifconfig:
sudo ifconfig ens7 mtu 8900 upto set the MTU to 8900. 8900 was (is?) the max MTU size for FABRIC in May 2022. I’m not sure if they have increased it since then. If you were at 9000, this would explain why you had a low throughput (dropped packets).I’m using
sudo tc qdisc add dev ens7 root fq maxrate 30gbitto use a fair-queuing model.And I write the following lines to
/etc/sysctl.conf(on Ubuntu 20 if that matters, I don’t think so…):# increase TCP max buffer size setable using setsockopt() net.core.rmem_max = 536870912 net.core.wmem_max = 536870912 # increase Linux autotuning TCP buffer limit net.ipv4.tcp_rmem = 4096 87380 536870912 net.ipv4.tcp_wmem = 4096 65536 536870912Hope this helps!
– Brandon
Hi Chengyi,
Something has definitely changed as I once achieved ~98 Gbps when running iPerf in parallel on the SALT-UTAH link. This was the only link I personally was ever able to get anywhere close to 100 Gbps.
I suspect that the increase in active users has a lot to do with it, as my previous test was on May 16, 2022. Unless there were changes to the backend configuration at SALT-UTAH that I do not know about @Paul?
Running the same notebook today yielded 28.435 Gbps (combined over 10 parallel streams). I cannot remember if I previously used Connect X6s or Basic NICs to achieve the 98Gbps, but both Connect X6s at SALT and UTAH were reserved today, so I had to run the test with Basic NICs. I’ll rerun the tests with Connect X6s once they become available again.
August 4, 2022 at 1:31 pm in reply to: SSH Command Requires Bastion Key to be Added or Needs SSH Config File? #2580Ahh, I see. Huh, that’s tricky then. Well, I guess I’ll just continue to manually type the
ssh -F /path/to/ssh_config -i /path/to/slice_key node@IP.Thanks!
August 4, 2022 at 12:28 pm in reply to: Request – Add Site Abbreviation to Portal’s “Resources” Tab #2577Thank you! Glad to help.
Brandon
August 4, 2022 at 12:27 pm in reply to: SSH Command Requires Bastion Key to be Added or Needs SSH Config File? #2576Also, somewhat related for others who might be stuck – with the new FABlib update, it seems that if you use a custom slice key, you must pass a
-i /path/to/custom_slice_keywith or without the-F /path/to/ssh_config.Maybe I just totally forgot I had to do this, but there it is in case others are trying to use custom keys.
August 4, 2022 at 12:21 pm in reply to: SSH Command Requires Bastion Key to be Added or Needs SSH Config File? #2575@Hussam Right, that makes sense. But…
@Paul If the new way of configuring FABlib is to run the configure_environment notebook, shouldn’t the ssh_config file now always get created at
~/work/fabric_config/ssh_config. I guess we would only have to worry about if users move the ssh_config file or decide to create the file themselves without the notebook, which would be a very rare case I would think.Point being that if the current SSH Command doesn’t work, we might as well replace it with a command that works in most cases. Or correct me if I’m wrong, maybe the current Command works for most people and I’m just not setup correctly?
Hi Manas,
I might be missing exactly what you meant, but are you wanting to reserve a slice without using a Jupyter Notebook, or are you more interested in running scripts and programs directly on a node itself using a terminal?
The former is relatively straightforward, and a written tutorial to install FABRIC and FABlib locally can be found here. NOTE: these libraries only help reserve nodes, they are not required to interface or connect with nodes.
The latter is something I have been working a lot on trying to make easier. Currently, you have two options to run programs on nodes: FABlib’s
node.execute()and locally SSHing into the node and then running the command directly. Would a tutorial on how to SSH into a node and run a script, all from a local terminal be helpful? Both cases have the issue that the node does not have a display, so no graphical information can be displayed by default.If you need graphical output, say to view a MatPlotLib plot, you can setup X11 forwarding on the node to stream the graphical output from the node to your local computer. We will make a video tutorial on how to do this soon!
-
This reply was modified 3 years, 7 months ago by
Brandon Rice.
August 3, 2022 at 9:27 pm in reply to: SSH Command Requires Bastion Key to be Added or Needs SSH Config File? #2565Okay, I actually found a third way to SSH in: specify the bastion key in a ProxyCommand argument:
ssh rocky@<IP> -oProxyCommand="ssh -W [%h]:%p <My Bastion Username>@bastion-1.fabric-testbed.net -i /home/fabric/work/fabric_config/fabric_bastion_key" -i /home/fabric/work/fabric_config/slice_keyAt least there are many options!
For those wondering how to embed video/media into forum posts, I believe this forum should be able to embed the following media types: https://wordpress.org/support/article/embeds/
All you have to do is paste the URL to the media without making it a link, i.e.
Here is the “Hello FABRIC” YouTube tutorial video:
https // www youtube com/watch?v=UG0U73XkTZE
I have removed the periods and colons from the URL above so it does not embed the video. Use the actual full, unmodified URL of the media you want to embed.
So for now, I am good. Thank you for your help!
If you’d like to add ipysheet to the default notebook container, I’m sure others might use it in the future. It is basically an ipywidget for creating and visualizing spreadsheets. For example, instead of the text wrapping/overflow when displaying the table of available resources, we could use an interactive spreadsheet that maintains the formatting like:
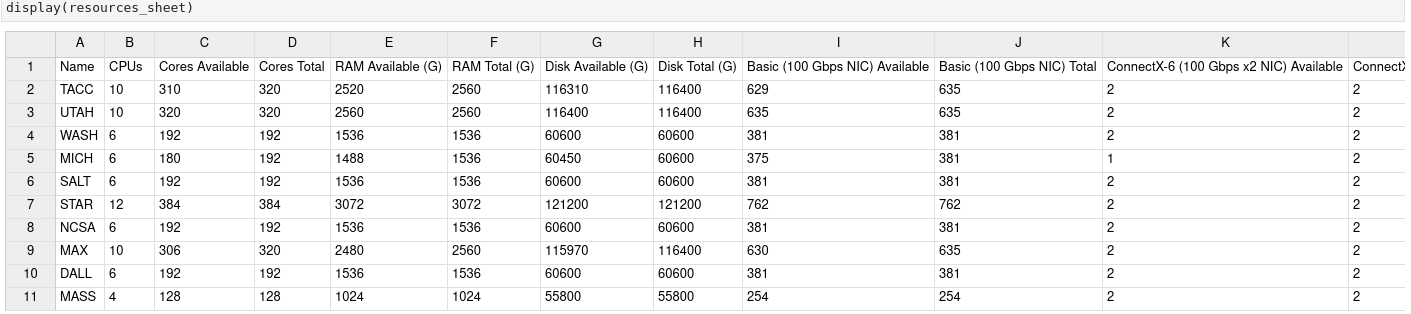
Hi Komal,
Thanks for looking into this. Following your steps to just copy the new files over the corrupted ones seems to work. However, by running a
conda update -y --all, this corrupted old json issue multiplies itself to all updated packages, and so if you try doing anything conda related after the conda update, you have this mess that it complains about (black highlighted files are corrupted):
Basically, just don’t do a conda update and it should work. I take it I could manually copy the newer versions over each of the corrupted files, but at this point my package that I originally wanted to install got installed successfully and works, so I am happy with the result and don’t need to do further digging right now. Hopefully the JupyterHub server resets itself so I don’t have to clean this mess up in the future.
Update: It seems the file exists but is corrupted or something?
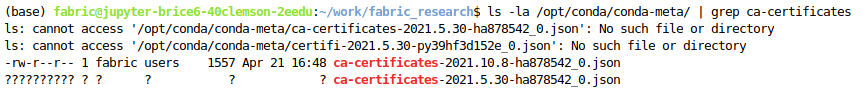
Any thoughts as to how to fix? Or maybe how to tell conda to use the one from 2021.10.8 that does exist normally?
-
This reply was modified 3 years, 7 months ago by
-
AuthorPosts
